Introduction
In a report published in 20181, the International Data Corporation (IDC) forecasts that the amount of generated data will grow from 33 Zettabytes (ZB) in 2018 to 175 ZB by 2025. To meet the demands, new storage media are being explored.
This article will be an introduction to DNA data storage, a promising alternative because its density and durability are superior to current silicon-based medium. No specific software knowledge is required to understand this topic, but you may need a refresher on your last biology course.
Some basic knowledge about DNA
The DNA (Deoxyribonucleic acid) is a molecule, present in each cell of an organism, that contains a genetic code. This code carries instructions for the development of the organism.
DNA is composed of pairs of nucleotides, of which there are four types:
- Adenine (A)
- Thymine (T)
- Cytosine (C)
- Guanine (G)
Writing a sequence of A, T, C and G pairs into actual DNA molecules is called synthesis. Reading the molecules back is called sequencing.
With these basic principles covered, you should easily understand the following article. Additional subjects requiring more background will be defined in the footnotes.
Key dates of DNA data storage
Let’s have a look at a non-exhaustive list of main works plublished on the topic.
1960: Basic concept of “molecular memory” systems is discussed by Norbert Wiener and Mikhail Neiman. DNA sequencing and synthesis technologies were in their early stages at that time, so the subject remained theoretical for 20 years.
1988: One of the first piece of data to be saved to DNA was actually an art
piece of 35 bit named “Microvenus” by Joe Davis.
He encoded an ancient germanic rune (𐌙) used to represent “female” and “life”.
Using their molecular weights, each of the four bases (C-T-A-G) where assigned
to phase-change2.
This language, one of many possible, was used to translate the binary image
into a sequence of base that was then synthesized into a DNA molecule.
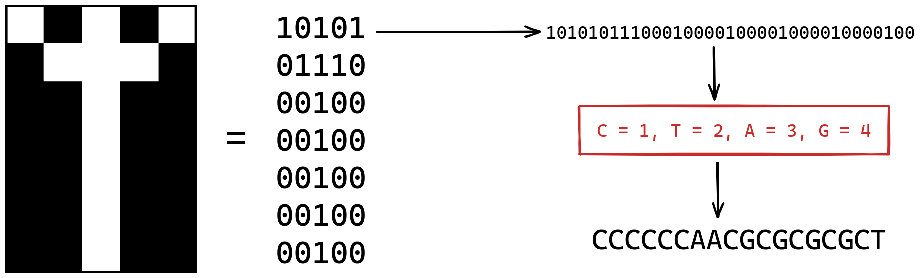
Fig. 1: The drawing of the rune is shown next to its binary representation. Using phase changes, the binary is translated to the four bases of DNA. The resulting strand of DNA is then synthesised and stored to be latter sequenced back to its binary format.
1999: Synthesized DNA was used to hide a “secret message” on paper with microdot3. The language used was a simple substitution cipher for the experiment but it was the first work, and remained the only until 2012, that did not include an in vivo step for the storage and the recovery process.
2012-2013: Two different teams4 revisited the topic of using DNA as a storage medium. Helped by the recent progress in writing and reading DNA, they both succeded at storing and reading back more than a kilobyte of information. After this, it became clear that DNA data storage could be viable and would have advantages over other storage methods.
Technique
Four major steps are involved in DNA data storage.
Write. This process starts by encoding the data by mapping strings of bits into DNA sequences using a translation algorithm. The resulting DNA sequences are then synthesized. Because those sequences are of finite length, bit strings have to be broken into smaller chunks, meaning we need a way to reassemble them into the original data. A simple index-based coding scheme is the most common solution to this problem.
Store. There are two possible ways to store the produced DNA material:
-
In vivo, which consist of cloning and storing the material within a biological cell. This was the most practical method with the knowledge of the early stages, but it offers an overall lower storage density due to the size of cells. The complexity of modifying or adding to natural DNA is another drawback of this storage method.
-
In vitro, where the DNA is stored frozen or dried down to achieve a better protection from the environment. This method offers better features, such as stability and scalibility, thanks to the controlled setting it provides.
Retrieve. Retrieving a data item on request is challenging in our context. It is made difficult by the lack of physical organization of the DNA strands in the same molecular pool. This issue is most often solved by using random access. It can be accomplished very easly as selective extraction of DNA fragments is common practice in molecular biology work.
The PCR-based5 approch is the most popular: during the writing process, the system assign “indexes” to the different chunks of data. Because we work with DNA, those “indexes” are unique primer pairs6 included in the corresponding DNA sequence. When a specific piece of data is requested, the DNA sequence containing the matching primers is amplified.
Read. The system sequences a sample of the resulting pool to retrieve the data. A set of reads is produced, corresponding to the molecules detected by the sequencer. The reads are then decoded back into the original format of data.
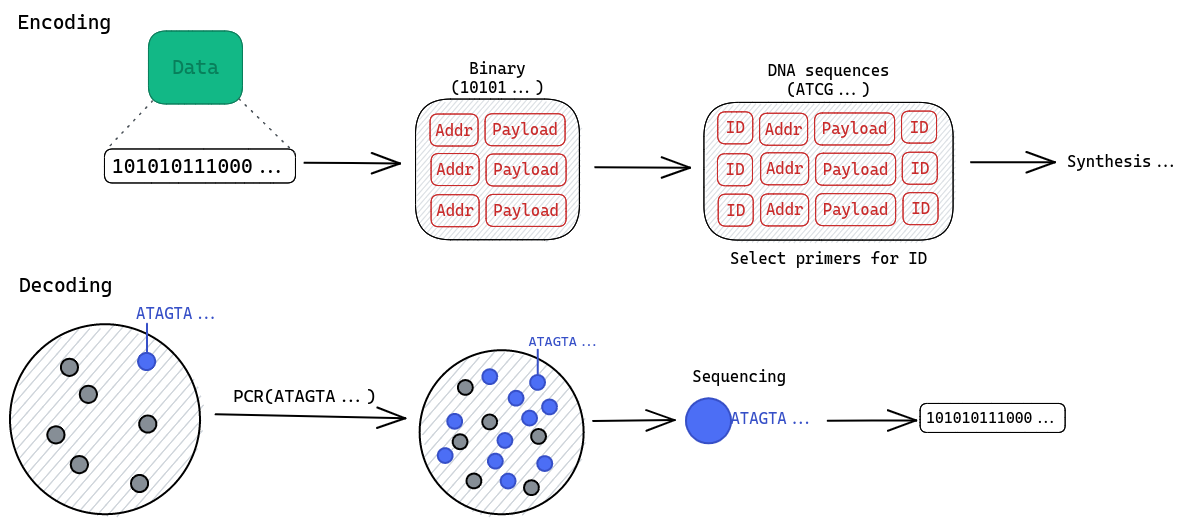
Fig. 2: Overview of the encoding-decoding process using the PCR approach.
How are errors handled during encoding/decoding
Like any data storage medium, DNA-based data storage needs to deliver data reliably despite noise. But unlike common storage channels, DNA, by its nature, can experience base insertions or deletions in addition to classic substitution errors. Several papers7 have reported an error rate of about 1% of the reads, after synthesis and sequencing.
The basis of error correction codes is always to add redundancy to the data, thus increasing the probability of correct recovery of the original information. Redundancy comes in two forms:
- Physical redundancy, since DNA synthesis naturally produces many physical copies of a given DNA sequence, this part is easily addressed.
- Logical redundancy is created by incorporating additional information during the encoding of data to DNA sequences.
A series of transformations and checks are passed by the bits we translate to DNA sequences. We have already seen that indices are added to identify the location of sequence in the original data. The use of a logical XOR operation with a seeded pseudo-random number is also widely used to ensure that each sequence is different.
More error correction codes can be used to add more redundancy. For example, the Reed-Solomon codes8 is commonly used for DNA-based storage. This method maps the original data into a set of symbols. These symbols are related to the coefficients of a system of linear equations, which solutions are mapped back to the original data.
Comparison with traditional storage mediums
Cons
Current data storage formats are now approaching their density limits. DNA is a promising alternative, but its mainstream adoption faces challenges.
Even with a DNA pool provisioned for PCR-based random access, the time to read is high (minutes to hours). In the short and medium terms, retrieving all data will stay a slow and steady process. Research is underway to create physicaly isolated pools that can be retrieved on demand without loss of density.
The writing throughput is estimated to be in the order of kilobytes per second, which is well below the Mb/s offered by more common mediums.
Last but not least, the cost needs to be reduced. The current cost gap between traditional, silicon-based storage and DNA storage is very high (source: Forbes, The Cost of Storage, 2016).
| Medium | $ per Gb |
|---|---|
| Tape | $0.02 |
| HDD | $0.033 |
| SSD | $0.25 |
| DNA | $800,000 |
The cost of DNA synthesis is often confidential, but Robert Carlson, notable industry analyst, estimated it to be $0.0001 per base9. Making it approximately $800k per gigabyte.
Pros
With its very slow read/write rate, DNA data storage cannot be used to replace our current HDDs or SSDs, it will not meet our needs. The main application area for this new medium is the archival data storage. For this type of application, higher latencies are tolerated for better durability and density.
DNA data storage is estimated to be 300 times more durable than the most stable magnetic tapes in an article of the Wyss Institute. Current storage medium are estimated to last a few decades before degrading, where DNA has a half-life10 of 500 years. Researchs are also conducted to auto-reproduce DNA. The DNA-coded data are placed into the genome of a very resistant microb, it can then reproduce itself at least a 100 times without data loss.
DNA synthesis, sequencing and retrieval technologies that are constantly developed for life sciences works can also be reused for DNA data storage. It is certain that those fields will continue to progress at the same time.
Conclusion
DNA data storage started early, with the first theories dating back to 1960. Nowadays, progress in molecular biology technologies made it clear that this new medium can coexist with commercial media. All four steps (writing, storing, retrieving and reading) are being actively researched to solve their bottlenecks.
In this article, we have taken a first dive into this ultra rich subject that touches many scientific fields. Some subjects were not covered, such as a detailed comparison of in vivo and in vitro storage. If you want to know more about these topics, go check out the bibliography, there will be articles and videos to peruse.
Bibliography
-
Ceze L, Nivala J, Strauss K. Molecular digital data storage using DNA. Nat Rev Genet. 2019 Aug;20(8):456-466. doi: 10.1038/s41576-019-0125-3. PMID: 31068682.
-
Davis, J. (1996). Microvenus. Art Journal, 55(1), 70–74. https://doi.org/10.2307/777811
-
Clelland, C. T., Risca, V., & Bancroft, C. (1999). Hiding messages in DNA microdots. Nature, 399(6736), 533–534. doi:10.1038/21092
-
Ted-Ed: Is DNA the future of data storage.
-
Microsoft Research: First fully automated DNA data storage.
-
The Digitization of the World, From Edge to Core by D. Reinsel, J. Gantz and J. Rydning, IDC and Seagate, 2018. ↩︎
-
How many times each binary bit is to be repeated before changing to the other binary bit. ↩︎
-
Type-written page photographed and greatly reduced to be pasted over a full stop in a letter so as to conceal messages. This was a widely used method by German spies during WWII. ↩︎
-
See Next-generation digital information storage in DNA (DOI) and Towards pratical, high-capacity, low-maintenance information storage in synthesized DNA (DOI) ↩︎
-
The Polymerase Chain Reaction is a commonly used method to rapidly make millions of copies of a specific DNA sample. In our case, the primers are used to amplify the sequences to which they are attached, thus allowing a detailed study. ↩︎
-
Primers are specific molecules in DNA who serves in the initiation of synthesizing strand of DNA. You can see it like the setup or init phase of program. ↩︎
-
See the section Coping with errors during encoding and decoding in the first entry of the bibliography for a list of studies on this matter. ↩︎
-
See Polynomial Codes Over Certain Finite Fields for more details (DOI). ↩︎
-
See Guesstimating the Size of the Global Array Synthesis Market (link) ↩︎
-
Half-life is the time it take for half the brands to break. ↩︎